Pandas Series

Creating pandas Series ¶
We are going to explore the pandas Series structure and learn how to store and manipulate single dimensional indexed data in the Series object.
The series is one of the core data structures in pandas. You think of it a cross between a list and a dictionary. The items are all stored in an order and there's labels with which you can retrieve them. An easy way to visualize this is two columns of data. The first is the special index, a lot like keys in a dictionary. While the second is your actual data. It's important to note that the data column has a label of its own and can be retrieved using the .name attribute. This is different than with dictionaries and is useful when it comes to merging multiple columns of data.
# Importing pandas
import pandas as pd
We can create a series by passing in a list of values. is, Pandas automatically assigns an index starting with zero and sets the name of the series to None. One of the easiest ways to create a series is to use an array-like object, like a list.
strings = ['a', 'b', 'c', 'd']
pd.Series(strings)
The result is a Series object which is nicely rendered to the screen. We see here that the pandas has automatically identified the type of data in this Series as "object" and set the dytpe parameter as appropriate. We see that the values are indexed with integers, starting at zero.
We don't have to use strings. If we passed in a list of whole numbers, for instance, we could see that panda sets the type to int64. Underneath panda stores series values in a typed array using the Numpy library. This offers significant speedup when processing data versus traditional python lists.
numbers = [4,2,3,9,0]
pd.Series(numbers)
[^top]
Missing data ¶
In Python, we have the None type to indicate a lack of data. If we create a list of strings and we have one element, a None type, pandas inserts it as a None and uses the type object for the underlying array.
strings = ['a', 'b', 'c', 'd', None]
pd.Series(strings)
If we create a list of numbers, integers or floats, and put in the None type, pandas automatically converts this to a special floating point value designated as NaN, which stands for "Not a Number".
numbers = [4,2,3,9,0,None]
pd.Series(numbers)
pandas represents NaN as a floating point number, and because integers can be typecast to floats, pandas went and converted our integers to floats. So when you're wondering why the list of integers you put into a Series is not floats, it's probably because there is some missing data.
NaN is not the same as None, though they are used to denote missing data.
import numpy as np
# Comparing NaN to None
np.nan == None
It would be interesting to note that two NaN values cannot be equated, so it's not possible to check if a value is NaN by using a conditional operator like ==.
np.nan == np.nan
Instead, we need to use special functions to test for the presence of not a number, such as the Numpy library isnan().
np.isnan(np.nan)
A series can be created directly from dictionary data. If you do this, the index is automatically assigned to the keys of the dictionary that you provided and not just incrementing integers.
# Here's a dictionary of fictional nations/kindgoms and their capital cities
capital_cities={'Azure':'Azure City',
'Land of Oz':'Emerald City',
'Discworld':'Ankh-Morpork',
'Carja':'Meridian',
'Kingdom of Loathing':'Seaside Town'}
cc=pd.Series(capital_cities)
cc
We see that, since it was string data, pandas set the data type of the series to "object". We see that the index, the first column, is also a list of strings.
We can get the index object using the index attribute.
cc.index
We can also separate your index creation from the data by passing in the index as a list explicitly to the series.
cc=pd.Series(['Azure City', 'Emerald City', 'Ankh-Morpork', 'Meridian', 'Seaside Town'],
index=['Azure', 'Land of Oz', 'Discworld', 'Carja', 'Kingdom of Loathing'])
cc
We can create a pandas series from a dictionary and also provide an index list. In this case, pandas will create a series with index from the index list instead of dictionay keys. It then matches the values in the index list with the dictionay keys. If there is a match, then the key-value pair are included in the series. If there is no match, then the value for that index is assigned as NaN.
pd.Series(capital_cities,index=['Land of Oz', 'Discworld', 'Carja', 'Coruscant'])
[^top]
Querying pandas Series ¶
A pandas Series can be queried either by the index position or the index label. If you don't give an index to the series when querying, the position and the label are effectively the same values. To query by numeric location, starting at zero, use the iloc[] attribute. To query by the index label, you can use the loc[] attribute.
If we wanted to see the fourth entry we would use the iloc[] attribute with the parameter 3.
cc.iloc[3]
We can query the same element using loc[] by specifying the index.
cc.loc['Carja']
Note that the indexing operators iloc and loc are not methods, but are attributes. So square brackets are used to query them instead of brackets.
In some cases, the usage of iloc[] and loc[] are optional as pandas automatically understands whether we are querying by index location or by the index name based on the input. This happens when the index values are not integers.
cc[3]
cc['Carja']
However, if the indices are numbers, omitting iloc[] or loc[] could be problematic.
# Prime numbers and their squares
prime_squares={2:4,3:9,5:25,7:49}
ps=pd.Series(prime_squares)
ps
Executing ps[0] would throw a key error because there's no item in the series with an index of zero.
ps[0]
It's a good practice to use iloc[] or loc[] explicitly to avoid such errors.
ps.iloc[0]
ps.loc[2]
[^top]
Pandas is fast and concise ¶
When working with data, we usually deal with performing operations of large set of numbers. This could be trying to find a certain number, or summarizing data or transforming the data in some way.
Let's generate a list of 10,000 random numbers and perform some basic operations ont them using lists and then with numpy and pandas and compare the time taken to perform these operations.
- We will first generate 10,000 random numbers.
- Perform a basic arithmetic operation of adding 5 to each.
- Calculate the average of the numbers.
We will time these operations using an iPython magic function called %%timeit. This function will run the code in a cell a specified number of times (1000 in this case) and calculate the average execution time. Cellular magic functions should be preceded with two percentage signs and should be on the first line of the cell.
import random
import numpy as np
import pandas as pd
Lists¶
# Generating a list of 10000 random numbers
rand_list=[]
for i in range(10000):
rand_list.append(random.random())
# Adding 5 to each number
for i in range(len(rand_list)):
rand_list[i]+=5
# Calculating the mean
rand_sum=0
for i in rand_list:
rand_sum+=i
rand_mean=rand_sum/len(rand_list)
print(rand_sum,rand_mean)
Now let's time it for an average of 1000 runs.
%%timeit -n 1000
# Generating a list of 10000 random numbers
rand_list=[]
for i in range(10000):
rand_list.append(random.random())
# Adding 5 to each number
for i in range(len(rand_list)):
rand_list[i]+=5
# Calculating the mean
rand_sum=0
for i in rand_list:
rand_sum+=i
rand_mean=rand_sum/len(rand_list)
It took about 15 milliseconds to perform the above operations using lists.
NumPy¶
Let's try doing the same with NumPy.
# Generating an array of 10000 random numbers
rand_array=np.random.random(10000)
# Adding 5 to each number
rand_array=rand_array+5
# Calculating the mean
rand_mean=rand_array.mean()
rand_mean
Let's time it for an average of 1000 runs.
%%timeit -n 1000
# Generating an array of 10000 random numbers
rand_array=np.random.random(10000)
# Adding 5 to each number
rand_array=rand_array+5
# Calculating the mean
rand_mean=rand_array.mean()
It took only 0.7 milliseconds with NumPy which about 20 times faster than with lists.
pandas¶
Let's try the same with pandas and see how fast it performs.
# Generating an array of 10000 random numbers
rand_series=pd.Series(np.random.random(10000))
# Adding 5 to each number
rand_series=rand_series+5
# Calculating the mean
rand_mean=rand_series.mean()
rand_mean
Timing it for 1000 runs
%%timeit -n 1000
# Generating an array of 10000 random numbers
rand_series=pd.Series(np.random.random(10000))
# Adding 5 to each number
rand_series=rand_series+5
# Calculating the mean
rand_mean=rand_series.mean()
It took 2 milliseconds to perform these operations using pandas Series. This is significantly faster than using lists. This is because pandas and the underlying numpy libraries support a method of computation called vectorization. Vectorization works with most of the functions in the numpy library, including the sum function. Put more simply, vectorization is the ability for a computer to execute multiple instructions at once, and with high performance chips, especially graphics cards, you can get dramatic speedups. Modern graphics cards can run thousands of instructions in parallel. We should start thinking about functional programming (as opposed to object-oriented programming) to utilise the power of parallel processing.
I was wondering why pandas Series was slower than NumPy arrays. Here is an explanation to it. While creating a pandas Series, pandas still depends on Python and makes calls to several Python functions. Here is a visualization of pandas indexing a series. Each coloured arc is a different function call in Python.
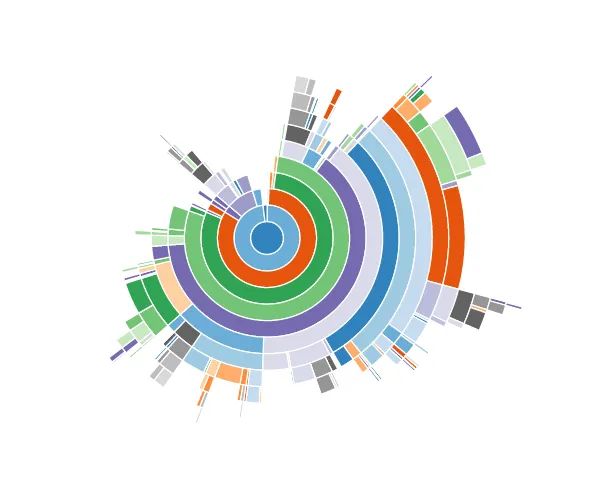
In contract, NumPy performs all the array indexing by itself and nothing is visible to Python. As you can see below, there are no Python function calls when using NumPy.
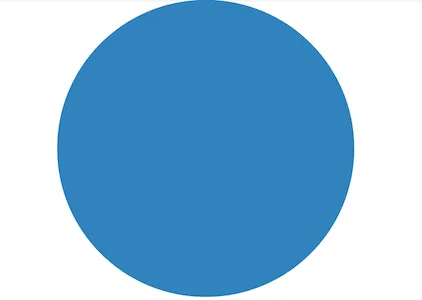
Another detailed comparision of execution times between pandas and NumPy is available here. According to the website, though NumPy is faster than pandas on smaller sets of data, pandas is equally fast and may be faster than NumPy as the size of the dataset grows. Pandas also offers a 2d dataframe structure and better usability than NumPy for data science operations.
[^top]
Manipulating pandas Series data ¶
The loc[] and iloc[] attributes lets us not only view data but modify data in place or add new data. If the value you pass in as the index doesn't exist, then a new entry is added.
# A series of 5 random numbers between 0 and 9 (inclusive)
x=pd.Series(np.random.randint(0,10,5))
x
Let's modify the third element and change it from 2 to 15.
x.iloc[2]=15
x
We can add a new element to the series using loc[].
x.loc['nine']=9
x
The indices need not be of the same data type but the series data must be of the same type. Pandas will type cast data to appropriate type. In this case, adding a floating point number changed all other integers to floating point numbers.
x.loc['decimal']=20.5
x
Adding a string converted the data type of the series to object.
x.loc['string']='zero'
x
Indices of pandas series are not unique like keys of a relational database.
y=pd.Series([0.9,9,9.9,99],index=['nine','nine','nine','nine'])
y
We can combine pandas series to a new series using append.
z=x.append(y)
z
When we query a pandas series by index, the result is all the values that match with that index.
z.loc['nine']
[^top]
Last updated 2020-12-20 17:49:22.201266 IST
Comments