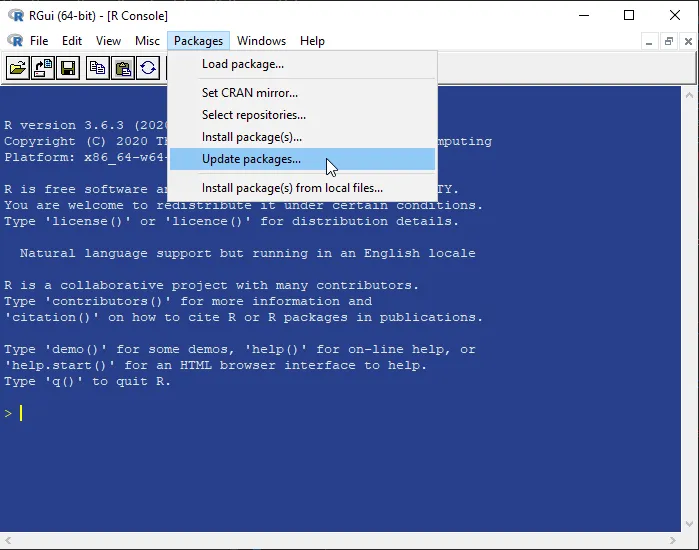
Vectors
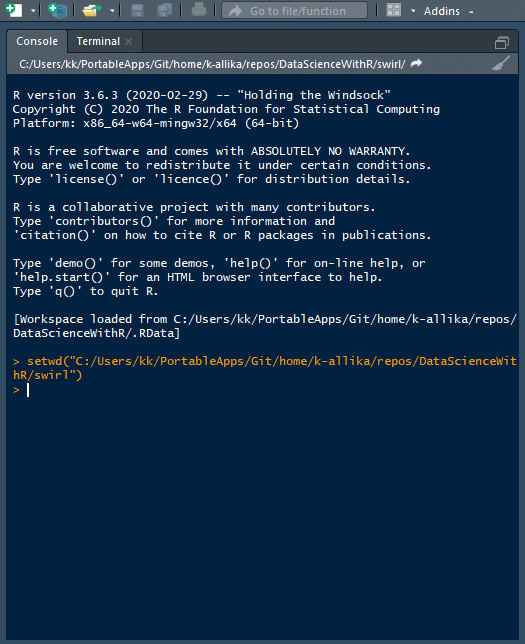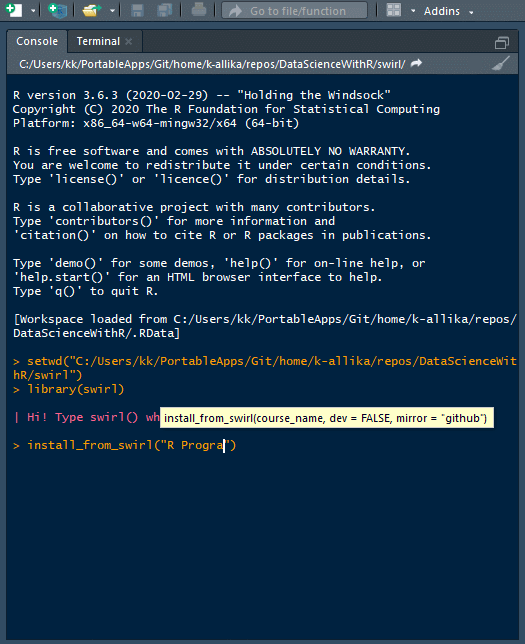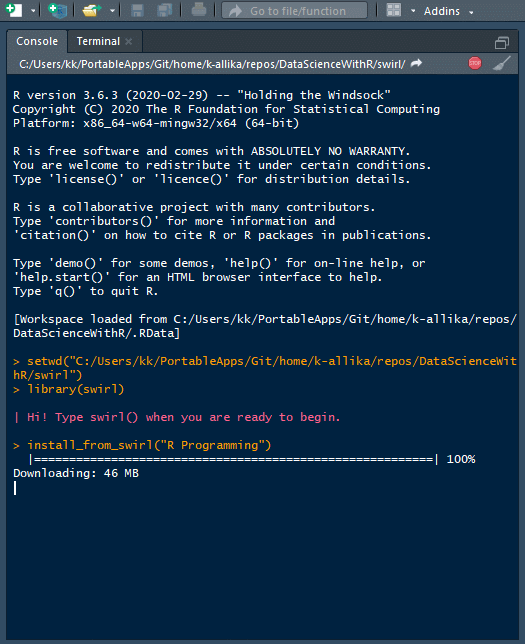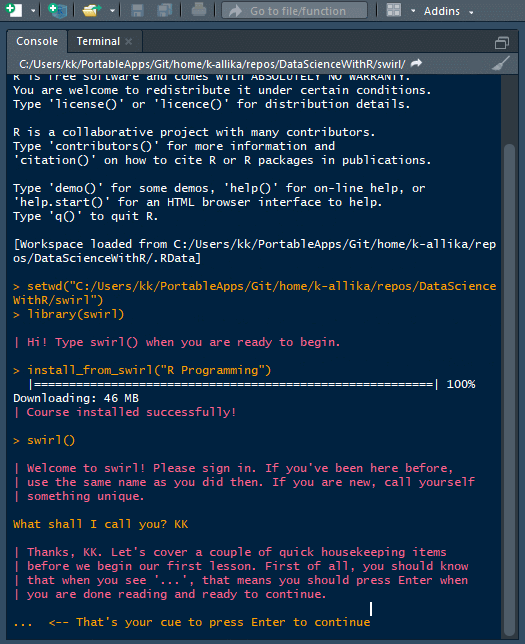
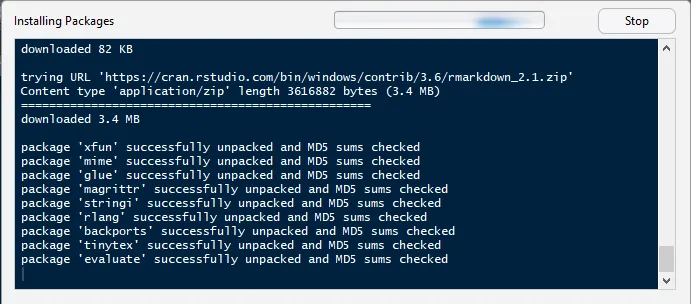
swirl()
| Welcome to swirl! Please sign in. If you've been here before, use the same name as you
| did then. If you are new, call yourself something unique.
What shall I call you? Krishnakanth Allika
| Please choose a course, or type 0 to exit swirl.
1: R Programming
2: Take me to the swirl course repository!
Selection: 1
| Please choose a lesson, or type 0 to return to course menu.
1: Basic Building Blocks 2: Workspace and Files 3: Sequences of Numbers
4: Vectors 5: Missing Values 6: Subsetting Vectors
7: Matrices and Data Frames 8: Logic 9: Functions
10: lapply and sapply 11: vapply and tapply 12: Looking at Data
13: Simulation 14: Dates and Times 15: Base Graphics
Selection: 4
| | 0%
| The simplest and most common data structure in R is the vector.
...
|== | 3%
| Vectors come in two different flavors: atomic vectors and lists. An atomic vector
| contains exactly one data type, whereas a list may contain multiple data types. We'll
| explore atomic vectors further before we get to lists.
...
|==== | 5%
| In previous lessons, we dealt entirely with numeric vectors, which are one type of
| atomic vector. Other types of atomic vectors include logical, character, integer, and
| complex. In this lesson, we'll take a closer look at logical and character vectors.
...
|====== | 8%
| Logical vectors can contain the values TRUE, FALSE, and NA (for 'not available'). These
| values are generated as the result of logical 'conditions'. Let's experiment with some
| simple conditions.
...
|======== | 11%
| First, create a numeric vector num_vect that contains the values 0.5, 55, -10, and 6.
num_vect<-c(0.5, 55, -10, 6)
| All that hard work is paying off!
|=========== | 13%
| Now, create a variable called tf that gets the result of num_vect < 1, which is read as
| 'num_vect is less than 1'.
tf<-num_vect<1
| You are really on a roll!
|============= | 16%
| What do you think tf will look like?
1: a vector of 4 logical values
2: a single logical value
Selection: 1
| Your dedication is inspiring!
|=============== | 18%
| Print the contents of tf now.
tf
[1] TRUE FALSE TRUE FALSE
| That's correct!
|================= | 21%
| The statement num_vect < 1 is a condition and tf tells us whether each corresponding
| element of our numeric vector num_vect satisfies this condition.
...
|=================== | 24%
| The first element of num_vect is 0.5, which is less than 1 and therefore the statement
| 0.5 < 1 is TRUE. The second element of num_vect is 55, which is greater than 1, so the
| statement 55 < 1 is FALSE. The same logic applies for the third and fourth elements.
...
|===================== | 26%
| Let's try another. Type num_vect >= 6 without assigning the result to a new variable.
num_vect>=6
[1] FALSE TRUE FALSE TRUE
| Great job!
|======================= | 29%
| This time, we are asking whether each individual element of num_vect is greater than OR
| equal to 6. Since only 55 and 6 are greater than or equal to 6, the second and fourth
| elements of the result are TRUE and the first and third elements are FALSE.
...
|========================= | 32%
| The < and >= symbols in these examples are called 'logical operators'. Other
| logical operators include >, <=, == for exact equality, and != for inequality.
...
|=========================== | 34%
| If we have two logical expressions, A and B, we can ask whether at least one is TRUE
| with A | B (logical 'or' a.k.a. 'union') or whether they are both TRUE with A & B
| (logical 'and' a.k.a. 'intersection'). Lastly, !A is the negation of A and is TRUE when
| A is FALSE and vice versa.
...
|============================= | 37%
| It's a good idea to spend some time playing around with various combinations of these
| logical operators until you get comfortable with their use. We'll do a few examples
| here to get you started.
...
|================================ | 39%
| Try your best to predict the result of each of the following statements. You can use
| pencil and paper to work them out if it's helpful. If you get stuck, just guess and
| you've got a 50% chance of getting the right answer!
...
|================================== | 42%
| (3 > 5) & (4 == 4)
1: FALSE
2: TRUE
Selection: 1
| That's correct!
|==================================== | 45%
| (TRUE == TRUE) | (TRUE == FALSE)
1: FALSE
2: TRUE
Selection: 2
| You are quite good my friend!
|====================================== | 47%
| ((111 >= 111) | !(TRUE)) & ((4 + 1) == 5)
1: FALSE
2: TRUE
Selection: 1
| Not exactly. Give it another go.
| This is a tricky one. Remember that the ! symbol negates whatever comes after it.
| There's also an 'order of operations' going on here. Conditions that are enclosed
| within parentheses should be evaluated first. Then, work your way outwards.
1: FALSE
2: TRUE
Selection: 2
| You nailed it! Good job!
|======================================== | 50%
| Don't worry if you found these to be tricky. They're supposed to be. Working with
| logical statements in R takes practice, but your efforts will be rewarded in future
| lessons (e.g. subsetting and control structures).
...
|========================================== | 53%
| Character vectors are also very common in R. Double quotes are used to distinguish
| character objects, as in the following example.
...
|============================================ | 55%
| Create a character vector that contains the following words: "My", "name", "is".
| Remember to enclose each word in its own set of double quotes, so that R knows they are
| character strings. Store the vector in a variable called my_char.
my_char<-c( "My", "name", "is")
| You're the best!
|============================================== | 58%
| Print the contents of my_char to see what it looks like.
my_char
[1] "My" "name" "is"
| Great job!
|================================================ | 61%
| Right now, my_char is a character vector of length 3. Let's say we want to join the
| elements of my_char together into one continuous character string (i.e. a character
| vector of length 1). We can do this using the paste() function.
...
|=================================================== | 63%
| Type paste(my_char, collapse = " ") now. Make sure there's a space between the double
| quotes in the collapse argument. You'll see why in a second.
paste(my_char,collapse = " ")
[1] "My name is"
| Great job!
|===================================================== | 66%
| The collapse argument to the paste() function tells R that when we join together the
| elements of the my_char character vector, we'd like to separate them with single
| spaces.
...
|======================================================= | 68%
| It seems that we're missing something.... Ah, yes! Your name!
...
|========================================================= | 71%
| To add (or 'concatenate') your name to the end of my_char, use the c() function like
| this: c(my_char, "your_name_here"). Place your name in double quotes where I've put
| "your_name_here". Try it now, storing the result in a new variable called my_name.
my_name<-c(my_char,"Krishnakanth Allika")
| You are doing so well!
|=========================================================== | 74%
| Take a look at the contents of my_name.
my_name
[1] "My" "name" "is"
[4] "Krishnakanth Allika"
| That's the answer I was looking for.
|============================================================= | 76%
| Now, use the paste() function once more to join the words in my_name together into a
| single character string. Don't forget to say collapse = " "!
paste(my_name,collapse = " ")
[1] "My name is Krishnakanth Allika"
| You got it!
|=============================================================== | 79%
| In this example, we used the paste() function to collapse the elements of a single
| character vector. paste() can also be used to join the elements of multiple character
| vectors.
...
|================================================================= | 82%
| In the simplest case, we can join two character vectors that are each of length 1 (i.e.
| join two words). Try paste("Hello", "world!", sep = " "), where the sep argument
| tells R that we want to separate the joined elements with a single space.
paste("Hello","world!",sep=" ")
[1] "Hello world!"
| Keep up the great work!
|=================================================================== | 84%
| For a slightly more complicated example, we can join two vectors, each of length 3. Use
| paste() to join the integer vector 1:3 with the character vector c("X", "Y", "Z"). This
| time, use sep = "" to leave no space between the joined elements.
paste(1:3,c("X", "Y", "Z"),sep="")
[1] "1X" "2Y" "3Z"
| Great job!
|===================================================================== | 87%
| What do you think will happen if our vectors are of different length? (Hint: we talked
| about this in a previous lesson.)
...
|======================================================================== | 89%
| Vector recycling! Try paste(LETTERS, 1:4, sep = "-"), where LETTERS is a predefined
| variable in R containing a character vector of all 26 letters in the English alphabet.
paste(LETTERS,1:4,sep="-")
[1] "A-1" "B-2" "C-3" "D-4" "E-1" "F-2" "G-3" "H-4" "I-1" "J-2" "K-3" "L-4" "M-1" "N-2"
[15] "O-3" "P-4" "Q-1" "R-2" "S-3" "T-4" "U-1" "V-2" "W-3" "X-4" "Y-1" "Z-2"
| You nailed it! Good job!
|========================================================================== | 92%
| Since the character vector LETTERS is longer than the numeric vector 1:4, R simply
| recycles, or repeats, 1:4 until it matches the length of LETTERS.
...
|============================================================================ | 95%
| Also worth noting is that the numeric vector 1:4 gets 'coerced' into a character vector
| by the paste() function.
...
|============================================================================== | 97%
| We'll discuss coercion in another lesson, but all it really means is that the numbers
| 1, 2, 3, and 4 in the output above are no longer numbers to R, but rather characters
| "1", "2", "3", and "4".
...
|================================================================================| 100%
| Would you like to receive credit for completing this course on Coursera.org?
1: Yes
2: No
Selection: 1
What is your email address? xxxxxx@xxxxxxxxxxxx
What is your assignment token? xXxXxxXXxXxxXXXx
Grade submission succeeded!
| You are doing so well!
| You've reached the end of this lesson! Returning to the main
| menu...
| Please choose a course, or type 0 to exit swirl.
Last updated 2020-10-01 18:12:28.577944 IST